What is Cross-Validation? Perform K-fold cross-validation without sklearn cross_val_score function on SVM and Random Forest
Cross-validation is a model evaluation method. Generally,
we use it to check the overfitting of the
trained model. If the whole dataset is divided into
train and test data, then the chances are high that the train model might not perform well on unseen (test) data. We can tackle
this problem by dividing the whole dataset into 3 sets: train, validation and
test dataset. Validation set will be used to check performance before test data
is applied. There are several methods to perform cross-validation such as holdout, K-fold, leave-one-out cross-validation.
First, we will import the required packages and load data. Here, we’ll be using iris data (link).
There are plenty of machine learning tools in python. The range
varies from sci-kit-learn, TensorFlow,
Keras, Theano, Microsoft CNTK. All of these provide excellent support and tools
to build your models, work through datasets. In this post, we will see an
alternate way to k-fold cross-validation.
Further, we will use the script for SVM and Random Forest Classifier.
K-Fold
Cross Validation
We divide the whole dataset into
k groups(folds) randomly, then we take one-fold as a test dataset and remaining
sets as training data. Then we train and
evaluate the model with that data. We repeat this procedure for k times. At last,
we take an average of all accuracy. It
mainly depends on how the data is divided in a random
way.
First, we will import the required packages and load data. Here, we’ll be using iris data (link).
from random import seed
import numpy as np
from random import randrange
iris_data = np.loadtxt('iris.data',delimiter=',')
Note that, randrange
and seed are to generate random sequence each
time.
Moving to the next step, in the cross_validation function
we are diving dataset as per fold size and taking data randomly from the
dataset for each fold. Once all the folds are ready with randomized data, we
will take one-fold as a test set.
def cross_validation(dataset, folds):
trainDataset = []
tempDataset = list(dataset)
# foldsize
foldSize = int(len(dataset) / folds)
for number in range(folds):
fold = []
while len(fold) < foldSize:
datasetIndexNumber = randrange(len(tempDataset))
fold.append(tempDataset.pop(datasetIndexNumber))
trainDataset.append(fold)
if len(dataset) % int(folds) == 0:
testDataset = trainDataset[-1:]
trainDataset.pop(-1)
return trainDataset, testDataset
Now, let’s prepare the model part where we can test our script.
We will be using SVM and Random Forest classifier for our model from sklearn package. You’ll have to download it to
your local environment if it is not installed.
First, we’ll transform our data (for each k values), so that we can feed it to our model and then we will train and predict
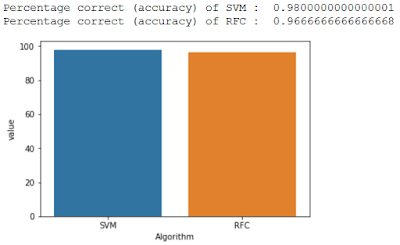
it for k times. Finally, we will take the mean from all iterations and plot the
accuracy of SVM and RFC.
seed()
count = 0
SVM_Values, RF_Values = [], []
cv = 10 # 10 fold cross validation
while count < cv:
# cross validation fucntion
trainDataset, testDataset = cross_validation(iris_data, cv)
# data transformation
tempTrainData = [item for sublist in trainDataset for item in sublist]
tempTestData = [item for sublist in testDataset for item in sublist]
trainDataset = np.array(tempTrainData)
testDataset = np.array(tempTestData)
# divide test and train set
x_train = trainDataset[:,0:-1]
y_train = trainDataset[:,-1]
x_test = testDataset[:,0:4]
y_test = testDataset[:,4]
# model
SVM_Model = svm.SVC(kernel='linear')
RF_Model = RandomForestClassifier(n_estimators=10)
# training the model
SVM_Model.fit(x_train,y_train)
RF_Model.fit(x_train,y_train)
# prediction
y_predicted_SVM = SVM_Model.predict(x_test)
y_predicted_RF = RF_Model.predict(x_test)
# storing predicted values for getting mean
SVM_Values.append(np.mean(y_test == y_predicted_SVM))
RF_Values.append(np.mean(y_test == y_predicted_RF))
count = count + 1
y_predicted_SVM_values = sum(SVM_Values) / cv
y_predicted_RF_values = sum(RF_Values) / cv
# plotting the graph
dataFrame = pd.DataFrame(data={'Algorithm': ['SVM','RFC'],
'Accuracy':[(y_predicted_SVM_values * 100), (y_predicted_RF_values * 100)]})
tempDataFrame = pd.melt(dataFrame,id_vars = ['Algorithm'],value_vars =['Accuracy'])
my_plot = sns.barplot(x="Algorithm", y="value", data=tempDataFrame)
#evaluation
print('Percentage correct (accuracy) of SVM : ', y_predicted_SVM_values)
print('Percentage correct (accuracy) of RFC : ', y_predicted_RF_values)
Comments
Post a Comment